Hello and welcome to The Anthony Lab website!
The Anthony Laboratory offers a dynamic environment at the intersection of nutrient sensing, metabolism, proteostasis, stress biology, and disease. We work with state-of-the-art molecular tools, animal models, omics approaches, and computational modeling to uncover how the body responds to nutritional challenges—and how these pathways can be leveraged to improve human health.
Lab’s Mission:
The mission of The Anthony Lab is to identify new ways to prevent and treat serious diseases and promote health span.
To achieve this we study metabolism and proteostasis in organ systems throughout the body. We are especially interested in sensing and signaling pathways that are responsive to nutrient scarcity and environmental stress. We also are interested in circadian and biological rhythms and the crosstalk between diet and physical activity.
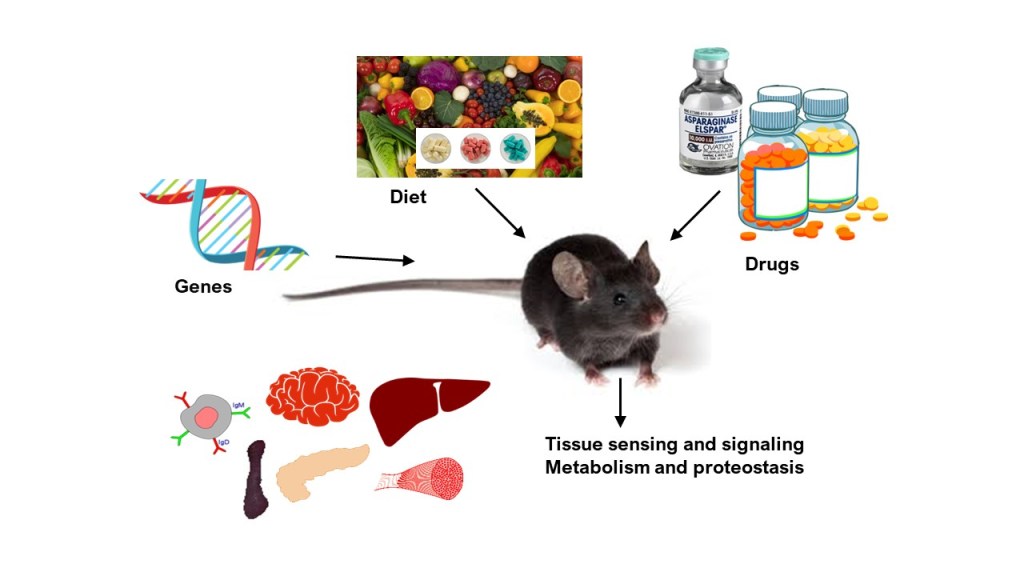
Research OVERVIEW
Our Big Question: How does the body sense nutrients and turn that information into decisions about growth, stress resistance, and survival?
The Anthony Lab studies how amino acids and other nutrients are detected at the cellular level, how this activates the Integrated Stress Response (ISR) and other related signaling pathways, and how those signals ripple out to affect metabolism, organ and tissue function, disease progression, and whole-body health.
We use a mix of mouse models, cell culture, omics (transcriptomics, proteomics, metabolomics), metabolic phenotyping, and computational modeling. We study nutrition at the molecular level all the way to the whole organism. Lab projects give trainees a strong conceptual grounding in how cells survive and adapt to stress.
Current projects in the laboratory may be grouped into the following areas:
click on the following titles below to learn more information
1. Amino Acid Sensing & the Integrated Stress Response
A core theme of the lab is understanding how cells detect amino acid insufficiency through the kinase GCN2, and how this triggers the ISR to collaborate with other signaling nodes such as the mechanistic target of rapamycin (mTORC1) to restore balance. While the mTORC1 pathway functions as a sensor of amino acid abundance to stimulate growth, the ISR is activated by amino acid insufficiency to slow growth. Simply put, mTORC1 functions as a gas pedal whereas GCN2 functions as a brake. Understanding how these signaling pathways work together to support cellular protection during stress is of great importance to supporting healthy aging and devising new and improved treatments for disease.
- We define how removal of specific amino acids (like leucine or sulfur amino acids) activates GCN2 and ISR programs in liver, reshaping translation and transcription.
- We show that GCN2 protects tissues especially liver from damage during amino acid depleting therapies like asparaginase, preventing oxidative stress, steatosis, and inflammation.
- We extend nutrient sensing into time: GCN2 is required for diurnal ISR activation and helps maintain circadian rhythms in energy expenditure, respiratory exchange ratio, and activity.
- We show that GCN2 is essential for nonshivering thermogenesis: mice lacking GCN2 cannot maintain core body temperature in acute cold and fail to appropriately uptake amino acids into brown adipose tissue.
- We were the first to describe negative regulation of mTORC1 by GCN2 and we continue to examine how the ISR regulates mTORC1 during different forms of cellular stress.
- We also continue to explore how the PERK-eIF2-ATF4 arm of the related Unfolded Protein Response contributes to the overall cellular effort to promote cellular adaptation and survival in response to a wide variety of environmental insults and proteotoxic stress agents.
For students: This area is ideal if you like molecular signaling, translation control, and a desire to learn how cells “measure” nutrient availability and translate that into changes in gene expression, metabolism, and organismal physiology.
2. Drug–Nutrient Interactions & Toxicities
Another major foci in the lab explores how clinically relevant drugs intersect with nutrient pathways. A key example is asparaginase, a bacterial enzyme used to treat acute lymphoblastic leukemia, the most common childhood cancer. Asparaginase breaks down circulating asparagine and glutamine, creating a systemic amino acid insufficiency. In leukemic cells which cannot make asparagine, asparaginase inflicts a lethal amino acid starvation. Yet for reasons not completely understood, cancer patients may unpredictably suffer severe complications, such as thromboembolism, liver failure and pancreatitis. These studies aim to increase the safety and efficacy of asparaginase and to develop new and improved uses for asparaginase to treat disease.
- We uncovered a bidirectional relationship between autophagy and the ISR in liver during asparaginase exposure, linking amino acid stress to food intake, energy expenditure, and liver health.
- We showed that the chemotherapy agent asparaginase doesn’t just deplete amino acids—it also disrupts vitamin A (retinoid) mobilization and liver retinol-binding protein trafficking, with implications for toxicity and tissue health.
- Systems-level studies, combining patient data records, metabolomics, and mouse experiments, point to retinoids as protective factors in asparaginase-associated pancreatitis, suggesting testable dietary or supplement strategies.
- We identified asparaginase as a specific activator of GCN2 and described the ISR as the body’s first responder to asparaginase, protecting the liver, pancreas and immune system during exposure.
- We also detailed the impact of obesity and age on liver responses to asparaginase exposure in mice.
- We continue to use this agent as a research tool to interrogate the nutrient sensing and signaling mechanisms that are triggered by amino acid insufficiency.
For students: This theme sits at the interface of nutrition, pharmacology, and precision medicine.
3. Dietary Amino Acid Restriction & Health span
The Anthony lab is a leader in studying dietary amino acid restriction as an intervention that improves metabolic health and may extend health span.
- Using kinetic proteomics, we showed how sulfur amino acid restriction rewires liver metabolism—altering lipid and amino acid handling, central carbon metabolism, and the ribo-interactome to support slowed growth but maintained proteostasis.
- We described that during sulfur amino acid restriction, brain protein synthesis is preserved while skeletal muscle synthesis is reduced. In contrast, genetic deletion of BCKDK (which enhances branched chain amino acid oxidation resulting in BCAA deficiency, epilepsy, and autism), reduced brain protein synthesis while skeletal muscle was preserved. These studies reveal a tissue hierarchy under nutrient stress that is amino acid specific.
- We used polysome profiling to reveal amino acid-specific regulation of ribosome activity and protein synthesis under nutrient scarcity.
- We study circadian and diurnal rhythms in liver metabolism during dietary amino acid insufficiency to better understand metabolic effects of shift work and jet lag.
For students: Projects here combine nutrition interventions with animal physiology, isotope labeling, omics, data science, and big-picture questions about metabolism at the ribosome.
4. Nutrient Stress in Cancer
The Anthony Lab collaborates closely with cancer biologists at Rutgers Cancer Institute of New Jersey and Indiana University School of Medicine to understand how tumors exploit nutrient stress pathways and how nutrient stress pathways affect cancer treatment strategies.
- In prostate cancer, we show that GCN2 is active and required for tumor growth by driving expression of amino acid transporters and maintaining intracellular amino acid pools. Pharmacologic GCN2 inhibition slows tumor growth in multiple models.
- We further reveal that GCN2 coordinates with p53 to support purine metabolism; disrupting both GCN2 and purine synthesis, or GCN2 and p53, pushes cancer cells from senescence toward cell death.
- In broader cancer and cachexia contexts, the lab contributes to understanding how nutrient stress, autophagy, and inflammatory signals drive wasting, appetite loss, and survival.
For students: This area combines cancer biology, metabolic vulnerabilities, and translation from mechanism to therapy.
5. Whole-Body Physiology : Metabolism in Motion
The lab doesn’t stop at signaling pathways—we connect them to whole-body function.
- In human and equine studies, we use metabolomics to map the “exercise metabolome,” in humans and horses, comparing exercise modalities and assessing how each modality perturbs and then restores circulating metabolites.
- In collaborative work, we reveal that loss of BCKDK reduces male fertility, revealing an underappreciated role for the branched chain amino acids (BCAAs) and especially the BCAA leucine in supporting testosterone synthesis and Leydig cell viability.
- The interface of dietary protein and exercise remains a longstanding area of interest and study. Previous projects include varying the composition, distribution, source and/or timing of dietary protein on mTORC1 signaling and protein synthesis. Information gained in this area will be targeted to relevant populations to improve performance and promote recovery and resilience.
For students: Projects here mix animal physiology, behavior, metabolic cages, and large datasets.

Sources of lab funding:


